|
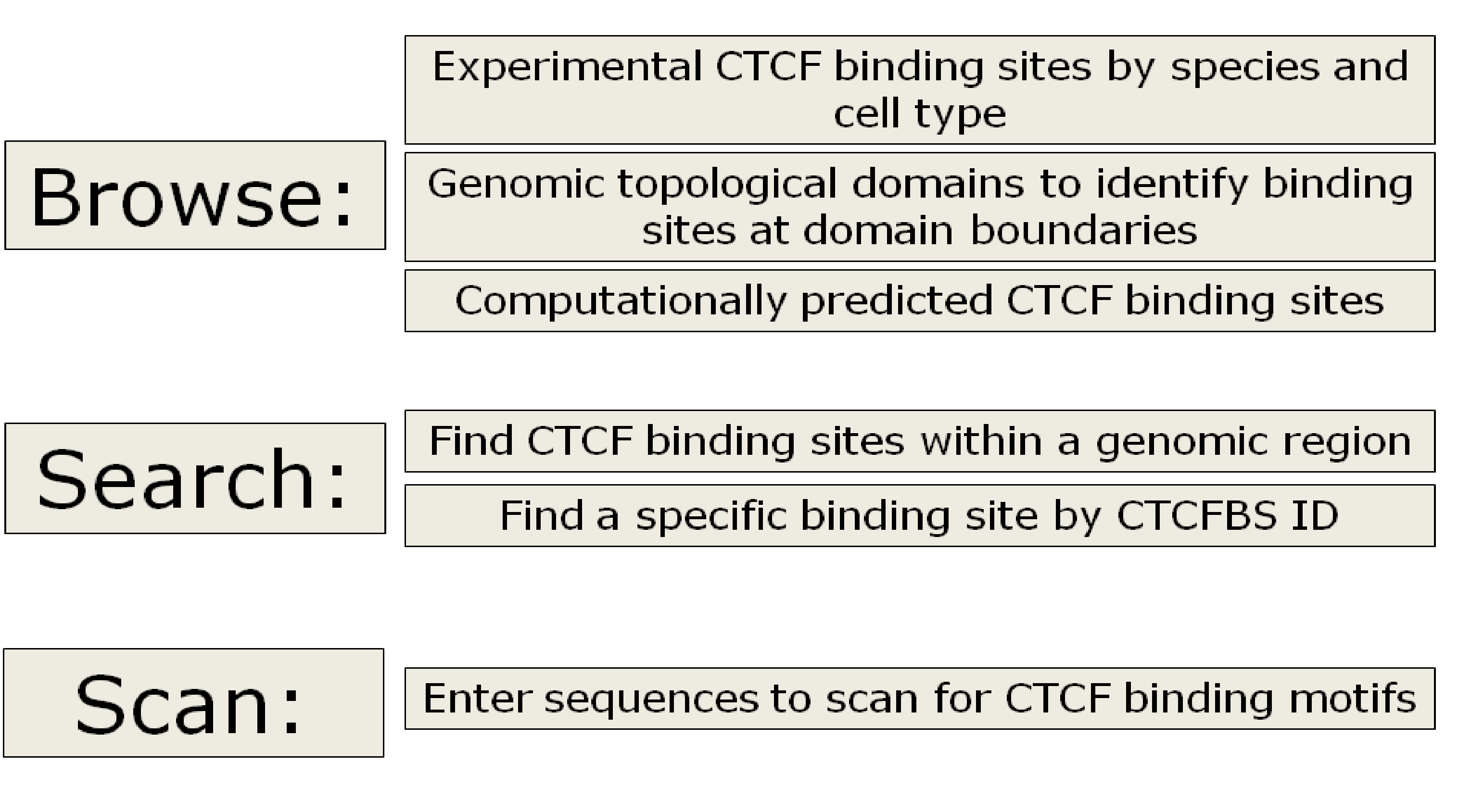
CCCTC-binding factor (CTCF) is a versatile transcription regulator that is evolutionarily conserved from fruit fly to human. CTCF binds to different DNA sequences through combinatorial use of 11-zinc fingers, and shows distinct functions (transcription activation/repression and chromatin insulation) depending on the biological context. Insulators, with the functions of enhancer-blocking and domain-bordering, are critical regulatory elements for gene expression control. They represent a class of diverged DNA sequences capable of shielding genes against inappropriate cis-regulatory signals from their genomic neighborhood. Recent studies also linked insulators to epigenetics, such as imprinting and X-chromosome inactivation. In eukaryotic genomes, maintenance of distinct chromatin domains is critical for transcription control, and CTCF has been identified as playing a crucial role in the global organization of chromatic architecture. Evidence for this CTCF function has been strengthened by Hi-C experiments that have shown that interacting genomic regions commonly contain CTCF binding sites and that the boundaries of genomic topological domains are enriched for CTCF binding sites. To analyze this important type of DNA regulatory element, we created a CTCF binding site database (CTCFBSDB), a comprehensive collection of experimentally determined and computationally predicted CTCF binding sites (CTCFBS) from the literature. The database is designed to facilitate the studies on insulators and their roles in demarcating functional genomic domains. Currently, the database contains almost 15 million experimentally determined CTCF binding sites across several species. CTCF binding sites were collected from published papers containing CTCF binding sites identified using ChIPSeq or similar methods, data from the ENCODE project, and a set of approximately 100 manually curated binding sites identified by low-throughput experiments. A complete list of the sources used to curate the CTCF binding sites within the database can be found on the Help page.
|